The White Spotting Series
Most white spotting on dogs is determined by the genes on the S locus. When we use the term “white spotting” we simply mean white areas on the dog, not actually white spots. White spotting can occur on any colour, and will cover up both eumelanin and phaeomelanin. In technical terms this is known as epistasis. So any dog can have white markings, whether theyre black, blue, liver, isabella, brindle, sable, tan-pointed, merle or whatever.
White hair occurs when the skin cells are unable to produce any pigment. The white spotting gene impairs the ability of cells on particular parts of the skin to make pigment, so the skin becomes pink and the fur white. Nails and paw pads will also become pink in areas where pigment is not produced.
So far, only two white alleles have been proven to exist on the S locus: S – no or very minor white sp – piebald
A third allele may exist for “extreme white” (sw), however this has not been proven and so far all dogs with high white have been shown to be homozygous for sp instead.
The white spotting alleles are thought to be examples of incomplete dominance. This means that a heterozygous dog will express its most dominant gene, but may also be affected by the more recessive one to a lesser extent. For example an Ssp dog may have some white spotting (see below). However, the relationship between the alleles is complicated and can vary between breeds.
It has been shown that some dogs with white spotting do not have an sp allele at all. These are mostly dogs with “true” irish spotting (in other words, irish spotting that breeds true – this should be made clear further down the page). The allele that causes this pattern has not yet been identified and it is not known if it is also located on the S locus. For the purposes of this site we will refer to this gene as si, but remember that it is most likely located on a different locus to sp. si is much less common than sp and only occurs in certain breeds.
Spread of White
Whichever white pattern a dog has, its white will always follow the same rules of spread. White starts on the farthest “edges” of the dog – the tail tip, the tip of the muzzle, the paws and the tip of the breastbone. This is known as the “trim” pattern. From there it spreads to cover the muzzle and forehead, the front of the chest, the lower legs and more of the tailtip, creating irish spotting. Next it spreads round from the front to the back of the neck, and creeps up the legs and tail. On a piebald dog, only the head, back and tail base may still be coloured. The back colouring is the next to go, followed by the tail base, then the face markings. The ears will always remain coloured unless the dog has a very high amount of white. The ears are generally the last part of the dog to turn white.
Of course the idea of white “spreading” is metaphorical, to give you a picture of how white patterning works. White doesnt spread like this on one particular dog (i.e. you wont get a solid coloured puppy that gradually loses colour as it grows, until its almost white! Although puppies do often lose or gain a little colour as they grow), its just to show which areas remain coloured on dogs with more and more white. One way to think of it is that the dog retains colour best in the most important areas of its body – around its internal organs (body and tail base patches) and its brain (ears and face patches) – and loses colour easiest from the parts farthest from these areas. In technical terms, pigment “migrates” to different parts of the body during the development of the embryo, and the S gene determines how far the pigment migrates. Sometimes it simply doesnt reach the furthest extremities (this can be caused by a minor problem or illness during development), and this can result in a small amount of white trim on a dog without sp, for example a small chest patch on an otherwise solid-coloured dog. Of course this means that technically, white doesnt spread at all – its actually the colour that spreads.
The white rules arent set in stone – sometimes individual dogs can have unusual white patterns, where, for example, the white on the legs is very uneven, or they have piebald patches in unexpected places, like on the neck or chest. However, in general, they do hold relatively true.
Residual White and White Trim
A very small amount of white on the chest, toes or tail may occur when the pigment doesnt migrate fully as the embryo develops. This is known as residual white and can sometimes be caused by minor illness in the mother or in the embryo, or may have no obvious cause at all. There is no particular genetic basis for residual white, however its clear that some dogs have a predisposition to it which can be passed on to their offspring.
If a slightly larger amount of white is present then the dog may be heterozygous for sp, in other words Ssp. In a breed such as the Newfoundland you would get such a dog from crossing a “Landseer” (piebald, spsp) with a solid (SS). However, in breeds carrying piebald there is no real way to know whether minimal white markings are just residual white or indicate the presence of the piebald gene without genetic testing or test breeding, as piebald heterozygotes may have anything from a tiny chest spot to pseudo-irish markings (see below).

 We can assume that the two dogs above are SS and that their markings are just residual white. This is because neither breed comes in piebald or irish spotting. If either of these dogs did have an sp or si gene then we would expect to see dogs with much more white being produced in these breeds, but this doesnt happen. As it is, their white is non-genetic and breeding two dogs with white markings in these breeds will not necessarily produce puppies with any white at all.
We can assume that the two dogs above are SS and that their markings are just residual white. This is because neither breed comes in piebald or irish spotting. If either of these dogs did have an sp or si gene then we would expect to see dogs with much more white being produced in these breeds, but this doesnt happen. As it is, their white is non-genetic and breeding two dogs with white markings in these breeds will not necessarily produce puppies with any white at all.
 This Staffordshire Bull Terrier is a possible piebald heterozygote (i.e. carrier of the piebald allele). We cant know for sure, but this is the most likely explanation for its white chest patch as the Staffie breed is known to commonly have the piebald gene. If this dog were bred to another sp carrier then some of the puppies may be piebalds and have much more white than either of their parents.
This Staffordshire Bull Terrier is a possible piebald heterozygote (i.e. carrier of the piebald allele). We cant know for sure, but this is the most likely explanation for its white chest patch as the Staffie breed is known to commonly have the piebald gene. If this dog were bred to another sp carrier then some of the puppies may be piebalds and have much more white than either of their parents.
Irish Spotting Pattern
Irish spotting (si) is the pattern sometimes known as “boston” or “mantle”, although these terms do not always refer to “true” irish spotting. On a dog with irish spotting, white is found on the legs, the tip of the tail, the chest, neck and muzzle. Many dogs with this pattern have a full white neck ring and a blaze.
True irish spotting is caused by an as yet unidentified gene, but we can assume irish spotted dogs to be homozygous for it (sisi) as it breeds true. This means that two irish spotted dogs bred together will produce puppies with irish spotting, and the white will not increase. We can assume that a solid dog bred to an irish spotted dog will produce a heterozygous dog with less white (a white trim, as shown in the section above).
The term “irish spotting” actually comes from a term used in the early 20th century to describe a white pattern found in rats in Ireland.



 The Aussies, Border Collie and Bernese Mountain Dog shown here are all true irish spotted. None of these breeds regularly come in piebald or extreme white and their white markings breed true (implying they are homozygotes).
The Aussies, Border Collie and Bernese Mountain Dog shown here are all true irish spotted. None of these breeds regularly come in piebald or extreme white and their white markings breed true (implying they are homozygotes).
Pseudo-Irish
“Pseudo” irish spotting may look the same or very similar to true irish spotting, but is in fact not caused by sisi but by Ssp, i.e. these dogs are really heterozyous or potentially even homozygous piebalds. The incomplete dominance of S means that an Ssp dog may show up to roughly half the amount of white as an spsp dog. These dogs do not breed true and when two are crossed the puppies may be solid, piebald or inbetween. See below for an example of this in Boxers.
Note that not all Ssp dogs show much white, or in some cases any white at all. The amount of white on a piebald heterozygote appears to vary drastically and some may look exactly like homozygous solids.



 The three breeds above (Staffie, Podengo Portugueso and English Pointer) all carry piebald but are not known to carry irish spotting, so these dogs are most likely pseudo-irish. A true irish spotted dog will not usually have white on the hips/knees or underside of the body, so this is another clue that sp is present.
The three breeds above (Staffie, Podengo Portugueso and English Pointer) all carry piebald but are not known to carry irish spotting, so these dogs are most likely pseudo-irish. A true irish spotted dog will not usually have white on the hips/knees or underside of the body, so this is another clue that sp is present.
Finally, a “flashy” irish spotted dog (one with more white than usual) may be caused by a combination of si and sp. If a true irish spotted dog also carries an sp allele then the normal white pattern may be extended. This supports the theory that si is actually on a different locus, as the two alleles appear to be inherited completely separately. This has been shown to occur in Shelties, where dogs carrying the sp allele as well as irish spotting can usually be identified by having more white around the neck and underside of the body. An spsp Sheltie has a high amount of white and is known as a “colour-headed white”. Shelties are one breed known to carry both true irish spotting and the sp allele, but many breeds only have one or the other.
 This irish spotted merle Sheltie has a large amount of white and may be a piebald carrier. Piebald carriers are often referred to as “white-factored” and are generally identified by having white extending further up the hind legs (onto the knees).
This irish spotted merle Sheltie has a large amount of white and may be a piebald carrier. Piebald carriers are often referred to as “white-factored” and are generally identified by having white extending further up the hind legs (onto the knees).
Piebald Pattern
Piebald (spsp) usually produces a coloured head (with or without white on the muzzle and as a blaze), and patches on the body. Generally the base of the tail is coloured, but other than that the patches may be located anywhere on the body (but rarely on the legs).
 Because piebald is a recessive gene and heterozygotes (piebald carriers) dont always have any white markings, it can remain hidden and pop up unexpectedly. Both the Poodle (as shown here) and the Shar Pei, traditionally solid-coloured breeds, occasionally produce piebald (known as “flowered” in Shar Pei).
Because piebald is a recessive gene and heterozygotes (piebald carriers) dont always have any white markings, it can remain hidden and pop up unexpectedly. Both the Poodle (as shown here) and the Shar Pei, traditionally solid-coloured breeds, occasionally produce piebald (known as “flowered” in Shar Pei).
Extreme White Pattern
The extreme white pattern consists of a completely or predominantly white dog with just small amounts of colour on its head and sometimes base of tail. Small body patches may be present too. Sometimes the nose is pink or partly pink, and the eyes may be blue in some breeds due to lack of pigment.
So far all extreme white dogs that have undergone genetic testing have been shown to be homozygous for the piebald gene (spsp), just like the piebalds in the section above. However, as there is a fairly large difference between those dogs and the ones shown below, it is possible there is something else going on to cause the high white. In breeds with both true irish spotting and piebald the high white may simply be caused by the interaction between homozygous irish spotting and homozygous piebald (e.g. the Sheltie). In other breeds the cause is less obvious and has led some people to postulate a further S allele – sw. However, no evidence has yet been found for the existence of sw, on the S locus at least.
Extreme white can occasionally cause problems when it removes large amounts of pigment from the face and ears. The most common problem is deafness (due to lack of pigment in certain parts of the inner ear, which prevents it from functioning properly), but dogs with exposed unpigmented (pink) skin are also more prone to skin cancer than those with more pigment.
Split Faces and White Heads
There is thought to be a separate gene or modifier that causes some dogs with the irish spotting, piebald or trim pattern to have a split or completely white face, even if theres very little other white on the dog. A split face is when half of the face is white and the other half is coloured.
Split and completely white faces are particularly common in the bull-type breeds, e.g. English Bulldog and Staffordshire Bull Terrier. The cause is unknown, however its interesting to note that many “panda” German Shepherds (see below), which are known to have a different white mutation to most other breeds, have split faces in a similar pattern, so its possible that split/white faces could be caused by the same or a similar gene.

 Saxon the Staffie submitted by Christina Jacobs (see Saxons Facebook page here), and Beagle submitted by Viktoria Kastner
Saxon the Staffie submitted by Christina Jacobs (see Saxons Facebook page here), and Beagle submitted by Viktoria Kastner
Ticking and Roan
Any white areas on a dog, no matter how big or small, may be ticked or roaned due to the T gene. The ticking corresponds to the colour the area of the coat would have been if it wasnt white, which means that it can vary across the body. For example, a black dog with tan points will have black/grey ticking where it would have been black, and red/tan ticking where the points would have been. See the Ticking page for more information.

 Australian Cattle Dog photo submitted by Viktoria Kastner Believe it or not, the Australian Cattle Dog above is an extreme white piebald. The solid black on the head is the actual markings, and the solid appearance of the rest of the coat is created by very heavy roaning, including the tan points. This dog will have been born white with colour on the head only, as roaning develops later. The Large Munsterlander to the right shows heavy ticking on a piebald dog. Ticking is generally lighter than roaning, and the individual spots may be larger.
Australian Cattle Dog photo submitted by Viktoria Kastner Believe it or not, the Australian Cattle Dog above is an extreme white piebald. The solid black on the head is the actual markings, and the solid appearance of the rest of the coat is created by very heavy roaning, including the tan points. This dog will have been born white with colour on the head only, as roaning develops later. The Large Munsterlander to the right shows heavy ticking on a piebald dog. Ticking is generally lighter than roaning, and the individual spots may be larger.
White Boxers
Boxers generally come in what appears to be the irish spotting pattern, so we would expect most examples of the breed to have sisi on the S locus. However, sometimes Boxer puppies are born which are completely or almost completely white. How these puppies could be regularly born to parents with much more colour perplexed Boxer breeders for a long time.
However, we can now provide an answer to this. Boxers do not have the si allele, and supposedly irish spotted Boxers are actually pseudo-irish – i.e. Ssp. When two pseudo-irish dogs are bred together some of the puppies will be homozygous piebalds (spsp). Boxers with no piebald allele can still have residual white on the extremities, and pseudo-irish dogs can have anything from a very small amount of white to full irish markings, which can sometimes make it difficult to distinguish a piebald carrier from a non-carrier.




 The illustrations above show the range of white markings in Boxers. It is likely that the sp allele here is affected by a modifier that extends the white markings, as white boxers are always extreme white instead of normal piebald pattern.
The illustrations above show the range of white markings in Boxers. It is likely that the sp allele here is affected by a modifier that extends the white markings, as white boxers are always extreme white instead of normal piebald pattern.
White puppies can therefore be mostly avoided by always breeding irish-marked dogs to solid dogs, although care must be taken when it is unclear whether a dog is genetically solid or pseudo-irish, as it is possible for solid dogs to carry a piebald allele and not express it at all or only partly.
White Boxers come with many of the problems associated with high-white dogs, including a high rate of deafness.
“Panda” Shepherds
So called “panda” German Shepherd Dogs, like the one pictured, have been found to have a completely different white mutation to normal dogs with this pattern. Although these dogs look like they have irish spotting, the pattern is actually caused by a different gene entirely, known as “KIT”. “Panda” is a dominant mutation, and like many white patterns caused by the KIT gene in other species, it is an embryonic lethal, which means that when an embryo has two copies of the gene it will be reabsorbed into the womb. Heterozygous dogs have no known health problems linked to the gene, however. Harlequin in Great Danes is another example of an embryonic lethal in dogs, although caused by a different gene to panda.
Some panda Shepherds have blue eyes, however this is not linked to the KIT gene. Another observation is that many panda Shepherds have split faces and very few seem to have the neat blazes often associated with true irish spotting or the piebald gene.
A few similar spontaneous white mutations in other breeds have been shown to have been caused by the same gene, including a mutation in Weimaraners. However, these mutations are very rare and the German Shepherd seems to be the only breed where a KIT mutation has become established.
“False” Whites
Sometimes white can occur on dogs separately to the S locus white spotting. One example is as part of the double merle pattern. A double merle will almost always have more white than its parents, and will often appear to have the piebald or extreme white pattern when in fact it does not carry those alleles. The harlequin gene also causes a similar effect. See the Merle page for more information.
White can also occur due to dilution of phaeomelanin by the I locus. Phaeomelanin is red pigment, and the I locus can dilute it to cream, ivory or sometimes even white. Breeds such as the Samoyed have this second type of dilution, so they appear completely white but in fact its not due to white spotting. They are recessive red (so they cannot produce any black pigment) with dilution of their red pigment to white, resulting in a solid white dog with black nose pigment. They also test as homozygous for recessive black, but it seems the recessive red overrides this.
The main way to tell a dog with extreme white spotting apart from a dog with phaeomelanin dilution is to look at the pigment on the nose, lips and eyerims. A dog with extreme white spotting is likely to be missing some pigment in these areas, so they will be partly or completely pink. A dog with phaeomelanin dilution will have solid black in all these areas (possibly with a dudley nose, which are common on dogs with dilution – see the Nose Colours page).

 These two dogs (a Siberian Husky and a Finnish Lapphund) are genetically black and tan (atat), but with dilution of their tan points to white, most likely to due the Northern domino (Ed) gene. It can be easy to mistake diluted points on a domino or black-and-tan dog for white markings, but points will generally be in a very regular and symmetrical pattern. The Husky has actual white spotting as well – note the irregular pattern on the chest and the thin blaze on the muzzle.
These two dogs (a Siberian Husky and a Finnish Lapphund) are genetically black and tan (atat), but with dilution of their tan points to white, most likely to due the Northern domino (Ed) gene. It can be easy to mistake diluted points on a domino or black-and-tan dog for white markings, but points will generally be in a very regular and symmetrical pattern. The Husky has actual white spotting as well – note the irregular pattern on the chest and the thin blaze on the muzzle.

 Urajiro is another phaeomelanin dilution pattern that can look like white markings. Urajiro is when the dilution is confined to the “points” of the dog, like on the Shiba Inu here. See the Phaeomelanin Dilution page for more information. Some dogs can have both phaeomelanin dilution and white spotting, like the Pembroke Welsh Corgi pictured above. This dog has the white collar associated with irish spotting, but also the symmetrical cheeks associated with urajiro. If you look closely, you can see that the cheeks are an “off-white” colour, not bright white like the collar.
Urajiro is another phaeomelanin dilution pattern that can look like white markings. Urajiro is when the dilution is confined to the “points” of the dog, like on the Shiba Inu here. See the Phaeomelanin Dilution page for more information. Some dogs can have both phaeomelanin dilution and white spotting, like the Pembroke Welsh Corgi pictured above. This dog has the white collar associated with irish spotting, but also the symmetrical cheeks associated with urajiro. If you look closely, you can see that the cheeks are an “off-white” colour, not bright white like the collar.
 First photo provided by Tina West, second by Dee Allison
First photo provided by Tina West, second by Dee Allison
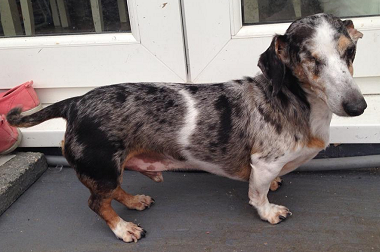
These two Dachshunds look just like piebalds, but theyre actually double merles and most likely dont have any S-locus white markings at all. The pattern produced by double merle is strikingly similar to a homozygous piebald, although it can sometimes be less regular and extreme split faces going all the way down the muzzle are very common (as on the second dog).




 One of these dogs is not like the others . . . but which is it? All are “false” whites except for one, which is an extreme white piebald. If you guessed the Staffie, youd be right. She has pink around her eyes, ears, muzzle and underside (a sign of lack of pigment, associated with extreme whites) and a few dark spots on her ears. All the other dogs are recessive reds (ee) or clear sables with phaeomelanin dilution. Note the slight cream sheen on the coat of the German Spitz, Samoyed and Shiba, and the jet black lip and eye rim pigment on all of them. The Shiba has a dudley nose, often associated with recessive red.
One of these dogs is not like the others . . . but which is it? All are “false” whites except for one, which is an extreme white piebald. If you guessed the Staffie, youd be right. She has pink around her eyes, ears, muzzle and underside (a sign of lack of pigment, associated with extreme whites) and a few dark spots on her ears. All the other dogs are recessive reds (ee) or clear sables with phaeomelanin dilution. Note the slight cream sheen on the coat of the German Spitz, Samoyed and Shiba, and the jet black lip and eye rim pigment on all of them. The Shiba has a dudley nose, often associated with recessive red.
|
Quick Summary! No time to read the whole thing? Heres the quick version! The S locus in dogs has two known alleles: S (no white markings), and sp (piebald). Due to incomplete dominance, one copy of the piebald allele results in a dog with minor white markings (often called the “trim” pattern), and two copies causes piebald or extreme white. A very wide range of patterns is caused by sp. In addition to sp, there is another allele known as si (irish spotting), which is most likely located on a different locus. A dog with one copy will have a white trim and a dog with two copies will have full irish spotting (white neck/collar, face, chest, legs and tail tip). The combination of si alleles and sp alleles is thought to cause most of the range of white markings found in dogs. Recently a rare mutation has been found in another gene known as KIT, and this causes the white markings on “panda” German Shepherd Dogs. |
|
Further Info and Links The gene causing the majority of white markings in dogs is known as MITF (Microphtalmia-Associated Transcription Factor). This gene causes white markings in a number of mammals and is often associated with blue eyes and deafness. The link between MITF and eye colour and hearing in dogs seems to be weaker than in some other species, although many high-white breeds such as Bull Terriers and Dalmatians do suffer from high rates of deafness. Interestingly, mutations in MITF have no link to skin colour in humans but do cause eye and sight issues. In other species, some white markings are also caused by the KIT gene. KIT is an extremely important gene and plays a role in stem cells and in the digestive tract, and some mutations in this gene (not those associated with white markings!) can also be associated with a number of different cancers. Most mutations in KIT causing white markings have no associated health problems when heterozygous, but are embryonic lethals when homozygous. KIT is responsible for “black-eyed white” phenotypes in many species, and has no association with either blue eyes or deafness. In dogs, KIT mutations have so far been confirmed in “panda” German Shepherds and in Weimaraners, but are not thought to be widespread. The other main gene causing white spotting in other species is EDNRB (Endothelin Receptor Type B). No mutations in this gene have been found in dogs, however in horses EDNRB causes “lethal whites” (overo lethal white syndrome), where homozygous foals do not have a fully functioning digestive system and die soon after birth. A number of human diseases are also associated with EDNRB. Links to studies: MITF and White Spotting in Dogs: A Population Study: http://jhered.oxfordjournals.org/content/100/suppl_1/S66.full A Simple Repeat Polymorphism in the MITF-M Promoter Is a Key Regulator of White Spotting in Dogs: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0104363 A de novo mutation in KIT causes white spotting in a subpopulation of German Shepherd dogs: http://www.ncbi.nlm.nih.gov/pubmed/23134432 Exclusion of EDNRB and KIT as the basis for white spotting in Border Collies: http://genomebiology.com/content/1/2/RESEARCH0004 Spotted Weimaraner: KIT Gene Strikes Again: http://colorgenetics.info/canine/spotted-weimaraner-kit-gene-strikes-again#sthash.XjGhqSlH.dpbs |
** Please note that I am not a research scientist, and the information on this page comes from my own knowledge and observation of dogs, observational and testing data provided via e-mail by site visitors, any research papers linked on the page, and the information provided by Dr Sheila M. Schmutz on her excellent website http://homepage.usask.ca/~schmutz/dogcolors.html
For further genetics resources, see the Links page
DNA is not as simple as ABC
The nuclei of dog cells contain important genetic data. The dog has 39 pairs of chromosomes in each cell (39 from the mother and 39 from the father). One of these pairs determines the sex of the dog and the rest determine everything else that makes him or her unique. Chromosomes are made up of thousands of genes that carry traits inscribed in DNA (see article “Genetic Basics: Understanding DNA” for more information).
Genes have pairs of alleles (one from each parent) that are located at specific sites (loci) on a chromosome. When dogs breed, the mother and father each randomly contribute one allele from each locus, giving each allele a 50% chance of being passed on to the pups. One of the alleles at each locus is dominant and determines the traits, like coat color, portrayed in the dog.
Despite the huge variety in coat color, there are only two basic pigments that determine the color of canines: eumelanin (black) and phaeomelanin (red). All different variations in color are created by these two pigments, which are both forms of melanin.
Melanocytes are the cells within the hair follicles that add melanin to the hair as it grows and determine basic coat color. The more melanin, the darker the color. Melanin is not always produced at a steady rate, so the tip of a dog’s hair may be darker than the rest of the hair shaft.
Each of the pigments, eumelanin and phaeomelanin, has a “default” color that can be modified by various genes. Eumelanin is, by default, black pigment, but variation in color occurs because genes modify eumelanin to create other colors such as liver (brown), blue (grey), or isabella (pale brown). Genes essentially “dilute” the pigment into these other colors by preventing the production of full strength eumelanin.
Phaeomelanin is the second pigment that determines canine coat color. This pigment is red with a default color of gold or yellow. Phaeomelanin creates reds that range from deep red (Irish Setter) to orange, cream, gold, yellow, or tan. Genes control the intensity of phaeomelanin, making the color stronger or weaker. This pigment is produced only in the coat and affects only hair color, while eumelanin affects eye and nose color. Phaeomelanin in people is responsible for freckles!
Eumelanin and phaeomelanin in all their forms create a huge range of dog coat colors. White hair on dogs occurs when cells do not produce any pigment at all. Most of the time this affects certain portions of the dog’s coat. For example, you may see a colored dog with white markings. If eumelanin is not produced in the nose, the dog ends up with a pink nose. If eumelanin is absent in the eyes, the dog has blue eyes. Rarely, the entire coat is affected, resulting in an albino dog with red eyes.
The fact that our domestic animals have a relatively long history (thousands of generations) and selection to change traits like coat color patterns has been very strong means that we now have a number of examples of the evolution of gene variants associated with several consecutive genetic alterations in the same gene and the MITF gene in dogs is one of the most beautiful examples of this, says Leif Andersson.
(Phys.org) —About half of all dogs show some form of white spotting which can range from a few white marks in the Bernese mountain dog to extreme white coat color in Dalmatians and white boxer. But why have dogs so often white markings, and how can we explain how they are determined genetically? This has researchers from Uppsala University, SLU and the Broad Institute spread new light on in an article that is now published in the scientific journal PLoS O
The Truth about the Merle Gene(SINE Insertion) in dogs
This study was designed to determine if one of the variants found in our laboratory, or previously reported in microphthalmia-associated transcription factor (MITF), was associated with one or more spotting patterns in dogs. None of the rare variants found in the coding sequence consistently occurred in dogs of any particular spotting pattern. However, an insertion of a short interspersed nucleotide element (SINE) over 3000 bp 5′ of the MITF-M start codon (Karlsson et al. 2007) did fit with random spotting in many dog breeds. Most (319/324) dogs of 45 breeds fit 1 of 2 inheritance patterns. All dogs that were homozygous for the SINE had white markings that either covered at least the ventral surface (mantle pattern) or most of the body (piebald or extreme white spotting). In most breeds, dogs heterozygous for the SINE insertion were solid colored or had minimal white, such as on the toes, but in some others, heterozygotes had white undersides, often with a white collar in the pattern called pseudo-Irish by Little (1957). However, none of the 15 dogs of 5 breeds in which all individuals have markings known as Irish spotting had the SINE insertion. Finally, we studied RNA expression in skin. The 2 MITF-M forms, M+ that contains an extra 18 bp that adds 6 amino acids between exons 5 and 6 and the M− form, were present. MITF-M is considered to be specific to melanocytes but was found in skin from a white Samoyed. A putative pseudogene containing exon 1M was also identified.
There are several patterns involving white markings in dogs. Little (1957) postulated that there was a single locus for white spotting in dogs, which he designated the S locus. He further suggested that 4 alleles explained the major patterns (Figure 1). S is the top dominant allele in the dominance hierarchy and causes solid or self-colored dogs. The next allele is si or “Irish spotting” that is a pattern in which there are white undersides, often a white neck collar, and sometimes white facial markings. The third allele is sp or piebald spotting in which random white spots occur on the body of the dog. The fourth allele is sw or extreme white in which the dog is almost entirely white but usually has at least some color on the head. Furthermore Little (1957) described a “pseudo-Irish” pattern that looks similar to the Irish spotting pattern but occurs in dogs that are S/sp heterozygotes (Figure 2, middle dog). Figure 1
Dogs representing the phenotypes caused by the 4 alleles postulated by Little (1957). Top left: S allele—solid German Longhaired Pointer. Top right: si—Shetland Sheepdog with Irish spotting. Bottom left: sp—piebald spotted Cocker Spaniel. Bottom right: sw—Japanese Chin with extreme white spotting. Figure 2
Italian Greyhound littermates (top row) and Collie parents and pup (bottom row) representing the codominant inheritance of spotting and their genotype for the SINE insertion 5′ of MITF. Left: solid or minimal white markings, center: pseudo-Irish pattern, right: extreme white with color head and minimal pigmented areas.
Little (1957) also discussed that plus and minus modifiers may explain extreme white as an extreme case of piebald and that on the other hand dogs with minimal white markings on the toes, for example, could still have an S/S genotype with minus modifiers. Pape (1990) also invoked plus and minus modifiers to explain the variation in piebald spotting in Newfoundlands.
Recently, 3 groups independently published that at least some forms of white spotting in dogs cosegregated with the gene microphthalmia-associated transcription factor (MITF) (Karlsson et al. 2007; Leegwater et al. 2007; Rothschild et al. 2006). Karlsson et al. (2007) and Leegwater et al. (2007) used a genome scan to map boxer patterns of white markings to CFA20, near MITF. Karlsson et al. (2007) then expanded their study to bull terriers to refine the chromosomal interval. They used haplotype analysis to suggest sets of variants present in some of the white spotting patterns. Rothschild et al. (2006) used a candidate gene approach and showed that families of Newfoundlands and crossbred dogs cosegregated for single nucleotide polymorphisms (SNPs) found in MITF and the presence or absence of piebald spotting.
In this study, we primarily focused on one variant in the 5′ untranslated region (UTR) reported by Karlsson et al. (2007). This variant is a short interspersed nucleotide element (SINE) insertion 3167 bp before the start codon of exon 1M (Supplementary Figure 1). We especially wanted to determine if this SINE insertion could be used as a marker to test for carriers of piebald spotting, which is considered to be recessive to solid color. We also tested its presence or absence in dogs with other spotting patterns. In addition, we studied litters of dogs that included either individuals that were or were not white spotted or that had different white spotting patterns to confirm that MITF variants cosegregated with these patterns.
We used cDNA prepared from skin biopsies, tail dockings, and surgeries collected from previous coat color studies (Schmutz et al. 2002) and placed in liquid nitrogen or RNAlater (Ambion) within 20 min of collection to obtain RNA sequence of MITF-M.
Cheek brush DNA samples (Epicentre, Madison, WI) from 12 dog families that segregated for white spotting patterns were obtained from dog breeders and owners and used for genotyping to confirm cosegregation with MITF variants. These included Chinese Shar-Pei (1), Collie (2), French Bulldog (2), “German Shepherd” (1), Great Dane (1), Icelandic Sheepdog (1), Italian Greyhound (2), and Newfoundland (2) families. Photographs were also supplied to document the pattern of white markings or lack thereof. In total, 324 individual dogs of 45 breeds were genotyped for the SINE insertion.
mRNA was extracted from skin biopsies as previously described (Berryere et al. 2003) using the TRIzol Reagent Isolation of Total RNA method (Gibco, Burlington, ON, Canada). The RNA was stored at −80 °C until cDNA preparation. RNA samples were digested with DNase I (Gibco), and then cDNA was prepared as previously described (Berryere et al. 2003) using Superscript Preamplification System for First Strand cDNA Synthesis (Gibco). The samples were stored at −80 °C.
Polymerase chain reaction (PCR) products greater than 1000 bp were resolved on a 1% agarose gel, whereas products less than 1000 bp were resolved on a 2% agarose gel, and the amplified bands were excised. Products were isolated using the QIAquick method (Qiagen, Mississauga, ON, Canada) and quantified on a 1% agarose gel using a DNA mass ladder. PCR fragments were sequenced at the National Research Council of Canada Plant Biotechnology Institute using an ABI Prism 373 Sequencer (Perkin Elmer Corporation) and the Big Dye Terminator kit (Perkin Elmer Corporation). Sequences were aligned using the Sequencher 4.8 computer software (Gene Codes Corporation, Ann Arbor, MI).
Various primers were designed for the purposes of sequencing MITF, as well as for detecting and isolating both MITF-M forms (Supplementary Table 1). Some of these primers were designed off of cattle MITF genomic sequence, prior to the release of the dog sequence. Others were designed off of dog sequence obtained in early sequence attempts using such primers or from later dog sequence in GenBank.
The PCR of 15 ml contained 50–100 ng of DNA template, 1.5 ml of 10× PCR buffer (Invitrogen, Carlsbad, CA), 1–3 mM MgCl2 (Invitrogen), 0.2 mM dNTP (Invitrogen), 0.5 U of Taq DNA polymerase (Invitrogen), 0.66 pmol of each primer, and 9.6 μl of ddH20. The PCR parameters consisted of an initial denaturation at 94 °C for 4 min, followed by 28 cycles of 50 s at 94 °C, 50 s at the appropriate annealing temperature for the primer pair (Table 1), and 50 s at 72 °C, followed by a final 4-min extension at 72 °C. To determine their relative amounts of both MITF-M forms, the products were resolved on a 4% DNA agar gel.
The microsatellite in the 5′ UTR was detected by using the appropriate primers in Supplementary Table 1 at 61 °C. A product of approximately 242 bp was run on a polyacrylamide gel to determine the alleles.
Karlsson et al. (2007) also discuss a length polymorphism in the 5′ UTR. We attempted to detect this by sequencing but were unable to reliably repeat our determination of the number of base pairs in the same individual. We also tried to run an amplified product that contained this region on a polyacrylamide gel and could not reliably show inheritance of it in families.
The complete mRNA sequence from the MITF-M+ isoform was obtained (GenBank NM_001003337). Skin from an Australian Shepherd was used to prepare the cDNA, and this breed is fixed for white undersides, typically known as Irish spotting.
MITF-M+ contains an additional 18 bp or 6 amino acids known as exon 6A (GenBank AY240952). We obtained both MITF-M+ and MITF-M− from cDNA prepared from pigmented dog skin from 2 Australian Shepherds and unpigmented skin of 1 Samoyed, in approximately equal proportions (Figure 3). It was assumed that the primers used would bind to both forms with equal opportunity, and therefore, the relative abundance of each form would be subject to relative abundance within the sample. Figure 3
Agarose gel showing a PCR product of MITF-M that spans exons 5 and 6 of mRNA prepared from dog skin. All 3 samples from different dogs (2 Australian Sheepdogs and 1 Samoyed) include the 18 bp known as exon 6A that distinguishes the M+ form, as well as the M− form in which it is absent.
Although we identified 2 variants in exonic sequence, neither affected the amino acid sequence. There is a c.771C>T variant in exon 8 and a c.*72A>G that is 72 bp past the stop codon in exon 9. None were consistently associated with any pattern of white markings.
Two microsatellites were detected in GenBank AAEX01006754 that contains MITF genomic sequence. One was a GAAA repeat 627 bp after the start of exon 1D and over 58 000 bp before exon 1M. It was heterozygous in some parents and therefore useful in those families to follow cosegregation. The other was a GT repeat detected in intron 6 that was not very polymorphic in the parents and hence did not yield further data.
An additional variant was discovered in a PCR product from genomic DNA that is very similar to MITF exon 1M (GenBank EU726637). However, it includes an SNP in the codon that would normally be the start codon (Supplementary Figure 2). Exon 1M is 158 bp, and there is a different nucleotide at 11 positions, and 4 codons are missing in the putative pseudogene. However, the exon 1M coding sequence is identical, except for a variant in the start codon. The sequence after exon 1M is much less homologous. There are different nucleotides at 15 of the first 80 bp, and then the homology is minimal (Supplementary Figure 2).
Because these 2 sequences could both be obtained from the same dogs, it was concluded that one is a pseudogene. The putative pseudogene sequence was obtained from 21 different dogs of 10 breeds, including dogs with and without white markings. Comparison of sequence obtained from mRNA from skin and genomic DNA from some of the same dogs made it possible to confirm which sequence was the actual exon 1M.
The location of this putative pseudogene has not been determined. Using basic local alignment search tool (BLAST) to the dog Build 2.1 sequence in the National Center for Biotechnology Information database, this segment matches 2 bacterial artificial chromosome sequences in GenBank (AC191512 and AC191629) well, but not perfectly.
Two Italian Greyhound families of 7 and 5 pups and 2 Collie families of 10 and 3 pups were studied, and both fit a codominant inheritance pattern as illustrated in Figure 2. Cosegregation with alleles at the microsatellite identified in the 5′ UTR and the SINE insertion and phenotype were found in the Italian Greyhounds (θ = 0.01, logarithm of the odds [LOD] = 2.96). Although all Collies have Irish spotting or white undersides, there were 2 families that showed additional white markings, also inherited in a codominant pattern that cosegregated with the SINE insertion. The Collie parents were homozygous for the microsatellites. The dogs heterozygous for the SINE had a much larger white neck collar, and the ins/ins homozygotes were mostly white, with pigmented heads and 1 or 2 pigmented body areas, a pattern called color headed in this breed (Figure 2) (θ = 0.01, LOD = 3.25).
One family of 8 Chinese Shar-Pei pups was solid or piebald spotted and cosegregated with the SINE insertion and the 5′ microsatellite. ins/ins Homozygotes were piebald, but both heterozygotes and the dogs without the SINE insertion were solid colored. Two families of 4 and 5 Newfoundland pups cosegregated for the solid and piebald spotting called Landseer in this breed. In this breed, some but not all heterozygotes had white hair on the toes and/or a white chest spot. All Landseer Newfoundlands in these families were homozygous for the SINE insertion.
Although German Shepherd Dogs do not typically have any white markings, there are some lines in which piebald markings occur. Both piebald, but none of the 4 pups nor 2 parents without white were SINE homozygotes, in one such family of 6 pups.
Two families of 5 French Bulldog pups cosegregated for solid and piebald spotting. Again all the piebald individuals were SINE homozygotes. One dog had a pattern known as mantle in some breeds (Figure 4), which is somewhat similar to Irish spotting, although the white markings cover the undersides and more of the thigh and torso. This dog was also a SINE homozygote. Figure 4
Three dogs with a pattern called mantle and their genotype for the SINE insertion 5′ of MITF. Left: French Bulldog, middle: Large Munsterlander, right: Great Dane. (This figure appears in color in the online version of Journal of Heredity.)
Mantle markings also occur commonly in Great Danes. Contrary to the French bulldog and 1 Large Munsterlander with markings that might be called mantle (Figure 4), 3 similarly marked Great Danes in a large family of 15 pups from 1 sire and 2 dams were heterozygous for the SINE, but another did not have a SINE.
Although most of the families cosegregated “perfectly” for a variant in MITF or a microsatellite near MITF and forms of white spotting, in one family of Icelandic Sheepdogs, the white marking phenotype did not cosegregate perfectly with the genotype at the SINE insertion for either dominant or codominant inheritance (Supplementary Figure 3). Note that the dam and sire of these 8 Icelandic Sheepdog pups were verified by parentage testing, even though one pups genotype of no SINE does not seem possible given the sires homozygous SINE insertion genotype.